About
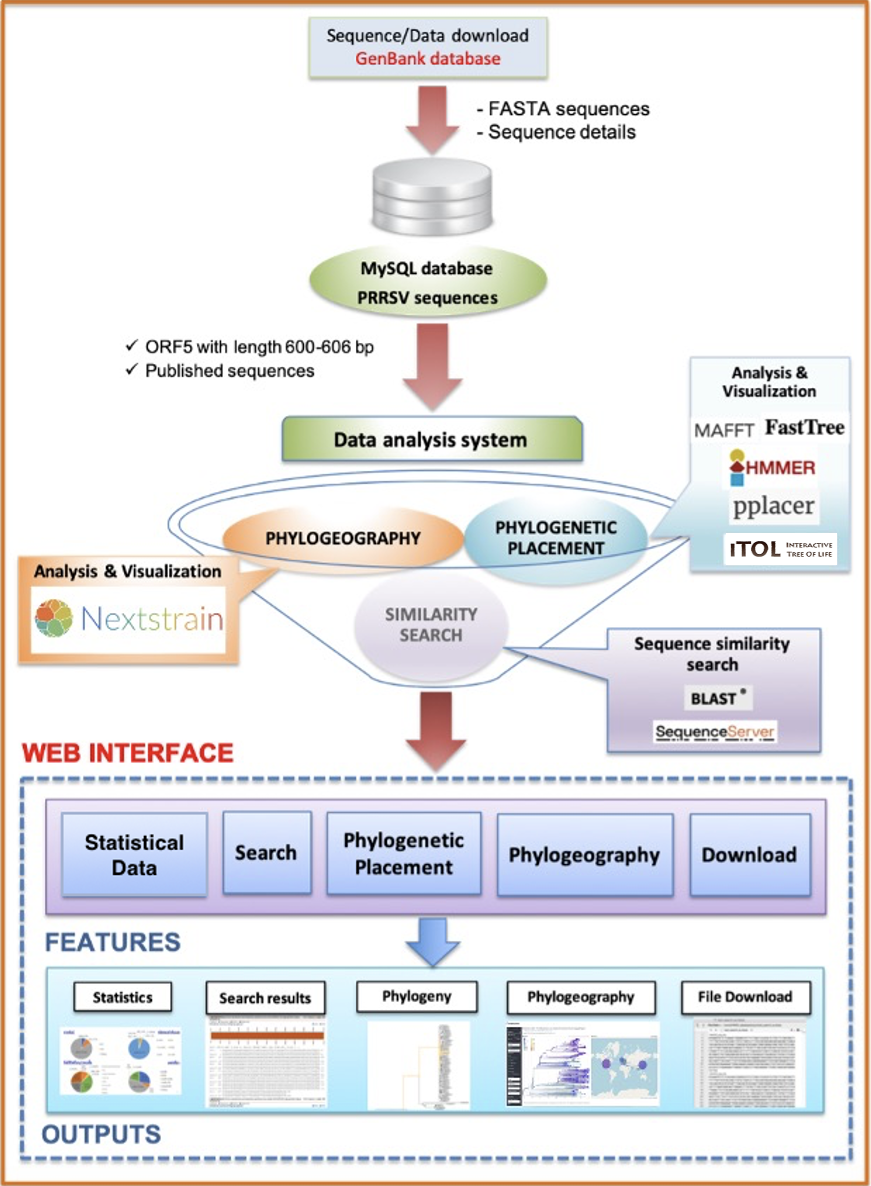
PRRSV-PDA currently includes 40,674 PRRSV sequences collected from GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) on Jan 18, 2024. The sequence data were also filtered to obtain the qualified sequences which were used as default dataset for all analyses. All data were categorized and summarized in the “Statistical data” section.
The current release of PRRSV-PDA consists of three data analysis modules.
1) Search :
Function: Allow sequence similarity search of ORF5 gene sequence between query and database sequences.
Required input: nucleotide sequence (s) in FASTA format.
Features:
- Select database by genotype and sequence origin.
- Can filter only the published sequences which were published in the specified time frame.
- Can include partially annotated and other sequences that have been filtered out before (e.g. sequences of unknown genotype/origin, unspecified gene sequences, unpublished sequences) as database sequences.
- Can set the search parameters.
Output: list of hit sequences, % query coverage, Evalue, % identity, query-match alignment.
2) Phylogenetic placement :
Function: Locate (Place) the query onto a fixed reference/backbone tree.
Required input: nucleotide sequence (s) in FASTA format or a FASTA file.
Features:
- Select a reference tree by continent.
- Tree visualization of the output tree.
Output: Reference tree with the placed query highlighted.
3) Phylogeograhy :
Function: Display pre-analyzed phylogeographic result of sequences obtained from specific continent(s).
Required input: No. Select only display parameters.
Features:
- Select analysis result by genotype and continent(s).
- Interactive visualization of the output.
Output: Animation of spatial phylogenetic processes through time.


